Single Point¶
janus-core contains various machine learnt interatomic potentials (MLIPs), including MACE based models (MACE-MP, MACE-OFF), CHGNet, and SevenNet.
Other will be added as their utility is proven beyond a specific material.
Set up environment (optional)¶
These steps are required for Google Colab, but may work on other systems too:
[1]:
# import locale
# locale.getpreferredencoding = lambda: "UTF-8"
# !python3 -m pip install janus-core[all]
Use data_tutorials to get the data required for this tutorial:
[2]:
from data_tutorials.data import get_data
get_data(
url="https://raw.githubusercontent.com/stfc/janus-tutorials/main/data/",
filename=["sucrose.xyz", "NaCl-set.xyz"],
folder="data",
)
try to download sucrose.xyz from https://raw.githubusercontent.com/stfc/janus-tutorials/main/data/ and save it in data/sucrose.xyz
saved in data/sucrose.xyz
try to download NaCl-set.xyz from https://raw.githubusercontent.com/stfc/janus-tutorials/main/data/ and save it in data/NaCl-set.xyz
saved in data/NaCl-set.xyz
Single point calculations for periodic system¶
[3]:
from ase.build import bulk
from janus_core.calculations.single_point import SinglePoint
# Change to cuda if you have a gpu or mps for apple silicon
device = "cpu"
NaCl = bulk("NaCl", "rocksalt", a=5.63, cubic=True)
sp = SinglePoint(
struct=NaCl,
arch="mace_mp",
device=device,
calc_kwargs={"model_paths": "small", "default_dtype": "float64"},
)
res_mace = sp.run()
NaCl = bulk("NaCl", "rocksalt", a=5.63, cubic=True)
sp = SinglePoint(
struct=NaCl,
arch="sevennet",
device=device,
)
res_sevennet = sp.run()
NaCl = bulk("NaCl", "rocksalt", a=5.63, cubic=True)
sp = SinglePoint(
struct=NaCl,
arch="chgnet",
device=device,
)
res_chgnet = sp.run()
print(f" MACE[eV]: {res_mace['energy']}")
print(f"SevenNet[eV]: {res_sevennet['energy']}")
print(f" CHGNeT[eV]: {res_chgnet['energy']}")
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/e3nn/o3/_wigner.py:10: FutureWarning: You are using `torch.load` with `weights_only=False` (the current default value), which uses the default pickle module implicitly. It is possible to construct malicious pickle data which will execute arbitrary code during unpickling (See https://github.com/pytorch/pytorch/blob/main/SECURITY.md#untrusted-models for more details). In a future release, the default value for `weights_only` will be flipped to `True`. This limits the functions that could be executed during unpickling. Arbitrary objects will no longer be allowed to be loaded via this mode unless they are explicitly allowlisted by the user via `torch.serialization.add_safe_globals`. We recommend you start setting `weights_only=True` for any use case where you don't have full control of the loaded file. Please open an issue on GitHub for any issues related to this experimental feature.
_Jd, _W3j_flat, _W3j_indices = torch.load(os.path.join(os.path.dirname(__file__), 'constants.pt'))
cuequivariance or cuequivariance_torch is not available. Cuequivariance acceleration will be disabled.
Downloading MACE model from 'https://github.com/ACEsuit/mace-mp/releases/download/mace_mp_0/2023-12-10-mace-128-L0_energy_epoch-249.model'
Cached MACE model to /home/runner/.cache/mace/20231210mace128L0_energy_epoch249model
Using Materials Project MACE for MACECalculator with /home/runner/.cache/mace/20231210mace128L0_energy_epoch249model
Using float64 for MACECalculator, which is slower but more accurate. Recommended for geometry optimization.
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/mace/calculators/mace.py:139: FutureWarning: You are using `torch.load` with `weights_only=False` (the current default value), which uses the default pickle module implicitly. It is possible to construct malicious pickle data which will execute arbitrary code during unpickling (See https://github.com/pytorch/pytorch/blob/main/SECURITY.md#untrusted-models for more details). In a future release, the default value for `weights_only` will be flipped to `True`. This limits the functions that could be executed during unpickling. Arbitrary objects will no longer be allowed to be loaded via this mode unless they are explicitly allowlisted by the user via `torch.serialization.add_safe_globals`. We recommend you start setting `weights_only=True` for any use case where you don't have full control of the loaded file. Please open an issue on GitHub for any issues related to this experimental feature.
torch.load(f=model_path, map_location=device)
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/nvidia_smi.py:810: SyntaxWarning: invalid escape sequence '\A'
mem = 'N\A'
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/nvidia_smi.py:831: SyntaxWarning: invalid escape sequence '\A'
maxMemoryUsage = 'N\A'
CHGNet v0.3.0 initialized with 412,525 parameters
CHGNet will run on cpu
MACE[eV]: -27.0300815358917
SevenNet[eV]: -27.057369232177734
CHGNeT[eV]: -29.32662010192871
Simple Molecules¶
[4]:
from ase.build import molecule
from ase.visualize import view
from janus_core.calculations.single_point import SinglePoint
sp = SinglePoint(
struct=molecule("H2O"),
arch="mace_off",
device=device,
calc_kwargs={"model_paths": "medium"},
)
res = sp.run()
print(res)
view(NaCl, viewer="x3d")
Downloading MACE model from 'https://github.com/ACEsuit/mace-off/raw/main/mace_off23/MACE-OFF23_medium.model?raw=true'
The model is distributed under the Academic Software License (ASL) license, see https://github.com/gabor1/ASL
To use the model you accept the terms of the license.
ASL is based on the Gnu Public License, but does not permit commercial use
Cached MACE model to /home/runner/.cache/mace/MACE-OFF23_medium.model
Using MACE-OFF23 MODEL for MACECalculator with /home/runner/.cache/mace/MACE-OFF23_medium.model
Using float64 for MACECalculator, which is slower but more accurate. Recommended for geometry optimization.
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/mace/calculators/mace.py:139: FutureWarning: You are using `torch.load` with `weights_only=False` (the current default value), which uses the default pickle module implicitly. It is possible to construct malicious pickle data which will execute arbitrary code during unpickling (See https://github.com/pytorch/pytorch/blob/main/SECURITY.md#untrusted-models for more details). In a future release, the default value for `weights_only` will be flipped to `True`. This limits the functions that could be executed during unpickling. Arbitrary objects will no longer be allowed to be loaded via this mode unless they are explicitly allowlisted by the user via `torch.serialization.add_safe_globals`. We recommend you start setting `weights_only=True` for any use case where you don't have full control of the loaded file. Please open an issue on GitHub for any issues related to this experimental feature.
torch.load(f=model_path, map_location=device)
{'energy': -2081.11639716893, 'forces': array([[ 0.00000000e+00, -3.88578059e-16, -6.47486826e-01],
[ 0.00000000e+00, -3.43765601e-01, 3.23743413e-01],
[ 0.00000000e+00, 3.43765601e-01, 3.23743413e-01]]), 'stress': array([0.00000000e+00, 6.12631596e-06, 4.50763482e-06, 2.43028903e-21,
0.00000000e+00, 0.00000000e+00])}
[4]:
Sugar on salt¶
[5]:
from ase.build import add_adsorbate, bulk
from ase.io import read, write
from ase.visualize import view
a = 5.63
NaCl = bulk(
"NaCl", crystalstructure="rocksalt", cubic=True, orthorhombic=True, a=5.63
) * (6, 6, 3)
NaCl.center(vacuum=20.0, axis=2)
sugar = read("data/sucrose.xyz")
add_adsorbate(slab=NaCl, adsorbate=sugar, height=4.5, position=(10, 10))
write("slab.xyz", NaCl)
sp = SinglePoint(
struct="slab.xyz",
arch="mace_mp",
device=device,
calc_kwargs={"model_paths": "small"},
)
res = sp.run()
print(res)
view(NaCl, viewer="x3d")
Using Materials Project MACE for MACECalculator with /home/runner/.cache/mace/20231210mace128L0_energy_epoch249model
Using float64 for MACECalculator, which is slower but more accurate. Recommended for geometry optimization.
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/ase/io/extxyz.py:311: UserWarning: Skipping unhashable information adsorbate_info
warnings.warn('Skipping unhashable information '
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/mace/calculators/mace.py:139: FutureWarning: You are using `torch.load` with `weights_only=False` (the current default value), which uses the default pickle module implicitly. It is possible to construct malicious pickle data which will execute arbitrary code during unpickling (See https://github.com/pytorch/pytorch/blob/main/SECURITY.md#untrusted-models for more details). In a future release, the default value for `weights_only` will be flipped to `True`. This limits the functions that could be executed during unpickling. Arbitrary objects will no longer be allowed to be loaded via this mode unless they are explicitly allowlisted by the user via `torch.serialization.add_safe_globals`. We recommend you start setting `weights_only=True` for any use case where you don't have full control of the loaded file. Please open an issue on GitHub for any issues related to this experimental feature.
torch.load(f=model_path, map_location=device)
{'energy': -1706.3958128381037, 'forces': array([[ 1.52358986e-09, 1.52359060e-09, 1.60311833e-01],
[ 5.89516217e-10, 5.89516399e-10, -2.49849858e-01],
[-3.71422142e-08, -4.76124738e-09, 2.55549362e-03],
...,
[-1.81414029e-01, 1.43043590e-01, -7.13535637e-02],
[ 1.37346116e-01, -1.33239451e-01, -1.05307659e-01],
[ 5.18585386e-02, 1.62407401e-01, -8.67263312e-02]]), 'stress': array([-4.33386986e-05, 7.45245471e-07, -1.69529958e-03, -5.32726537e-05,
-3.41297966e-05, -4.53043161e-05])}
[5]:
Calculate an entire collection of data frames¶
[6]:
from ase.io import read
import matplotlib.pyplot as plt
import numpy as np
frames = read("data/NaCl-set.xyz", format="extxyz", index=":")
dft_energy = np.array([s.info["dft_energy"] / len(s) for s in frames])
sp = SinglePoint(
struct="data/NaCl-set.xyz",
arch="mace_mp",
device=device,
calc_kwargs={"model_paths": "small"},
)
sp.run()
mace_mp_energy = np.array([s.info["mace_mp_energy"] / len(s) for s in sp.struct])
rmse_mace = np.linalg.norm(mace_mp_energy - dft_energy) / np.sqrt(len(dft_energy))
sp = SinglePoint(struct="data/NaCl-set.xyz", arch="chgnet", device=device)
sp.run()
chgnet_energy = np.array([s.info["chgnet_energy"] / len(s) for s in sp.struct])
rmse_chgnet = np.linalg.norm(chgnet_energy - dft_energy) / np.sqrt(len(dft_energy))
sp = SinglePoint(struct="data/NaCl-set.xyz", arch="sevennet", device=device)
sp.run()
sevennet_energy = np.array([s.info["sevennet_energy"] / len(s) for s in sp.struct])
rmse_sevennet = np.linalg.norm(sevennet_energy - dft_energy) / np.sqrt(len(dft_energy))
print(
f"rmse: mace_mp = {rmse_mace}, chgnet = {rmse_chgnet}, "
f"sevennet = {rmse_sevennet} eV/atom"
)
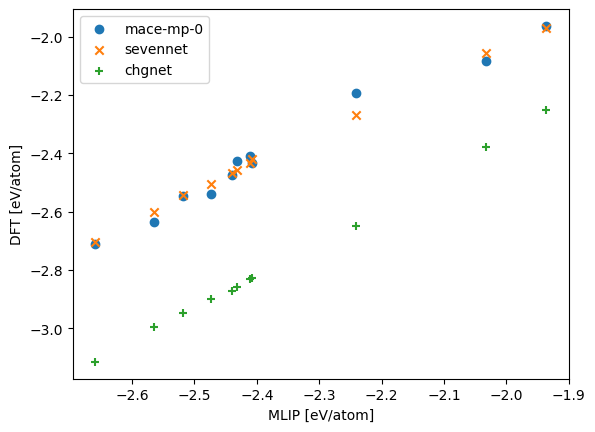
fig, ax = plt.subplots()
ax.scatter(dft_energy, mace_mp_energy, marker="o", label="mace-mp-0")
ax.scatter(dft_energy, sevennet_energy, marker="x", label="sevennet")
ax.scatter(dft_energy, chgnet_energy, marker="+", label="chgnet")
ax.legend()
plt.xlabel("MLIP [eV/atom]")
plt.ylabel("DFT [eV/atom]")
plt.show()
Using Materials Project MACE for MACECalculator with /home/runner/.cache/mace/20231210mace128L0_energy_epoch249model
Using float64 for MACECalculator, which is slower but more accurate. Recommended for geometry optimization.
/home/runner/work/janus-core/janus-core/.venv/lib/python3.12/site-packages/mace/calculators/mace.py:139: FutureWarning: You are using `torch.load` with `weights_only=False` (the current default value), which uses the default pickle module implicitly. It is possible to construct malicious pickle data which will execute arbitrary code during unpickling (See https://github.com/pytorch/pytorch/blob/main/SECURITY.md#untrusted-models for more details). In a future release, the default value for `weights_only` will be flipped to `True`. This limits the functions that could be executed during unpickling. Arbitrary objects will no longer be allowed to be loaded via this mode unless they are explicitly allowlisted by the user via `torch.serialization.add_safe_globals`. We recommend you start setting `weights_only=True` for any use case where you don't have full control of the loaded file. Please open an issue on GitHub for any issues related to this experimental feature.
torch.load(f=model_path, map_location=device)
CHGNet v0.3.0 initialized with 412,525 parameters
CHGNet will run on cpu
rmse: mace_mp = 0.04345175020411889, chgnet = 0.41196170963560735, sevennet = 0.02978418682071067 eV/atom